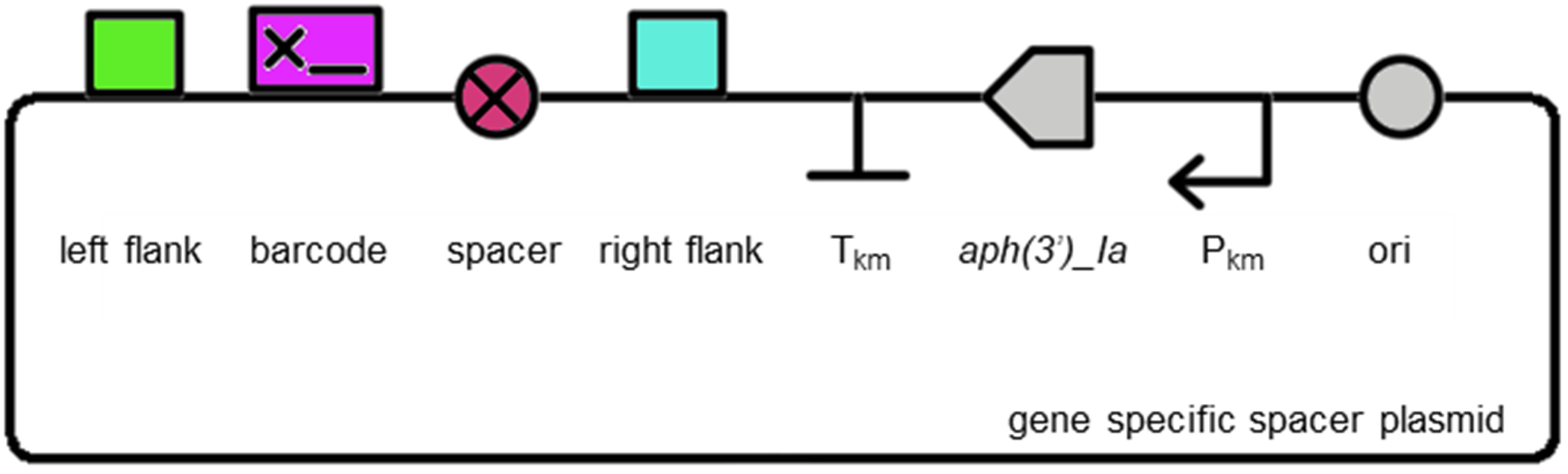
Gene specific knockout plasmids
Below is an example of a transformation plasmid assembled by MoClo assembly. Plasmids were assembled at the Foundries with a standard BsaI MoClo assembly protocol using 4 parts: 1) a pUC19 acceptor plasmid (selection with ampicillin), 2) a 'Left Flank' sequence homologous to the region upstream of the target gene (green), 3) a unique 18 bp barcode (purple) and an insertion sequence for selection and counter selection in Synechocystis sp. PCC 6803 (see below), and 4) a 'Right Flank' sequence homologous to the region downstream of the target gene (cyan). Type II restriction sites to facilitate additional editing are highlighted. These plasmids were designed to generate a marked gene knockout mutant by homologous recombination with the two flank sequences (see Overview).
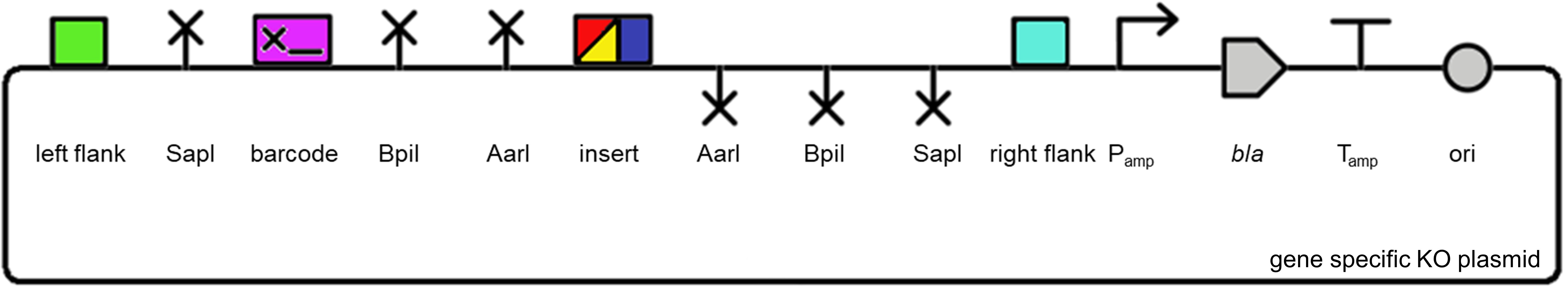
Insertion sequence
The insertion sequence consists of the kanamycin resistance cassette (KmR) for positive selection in Synechocystis sp. PCC 6803 to generate a marked mutant:
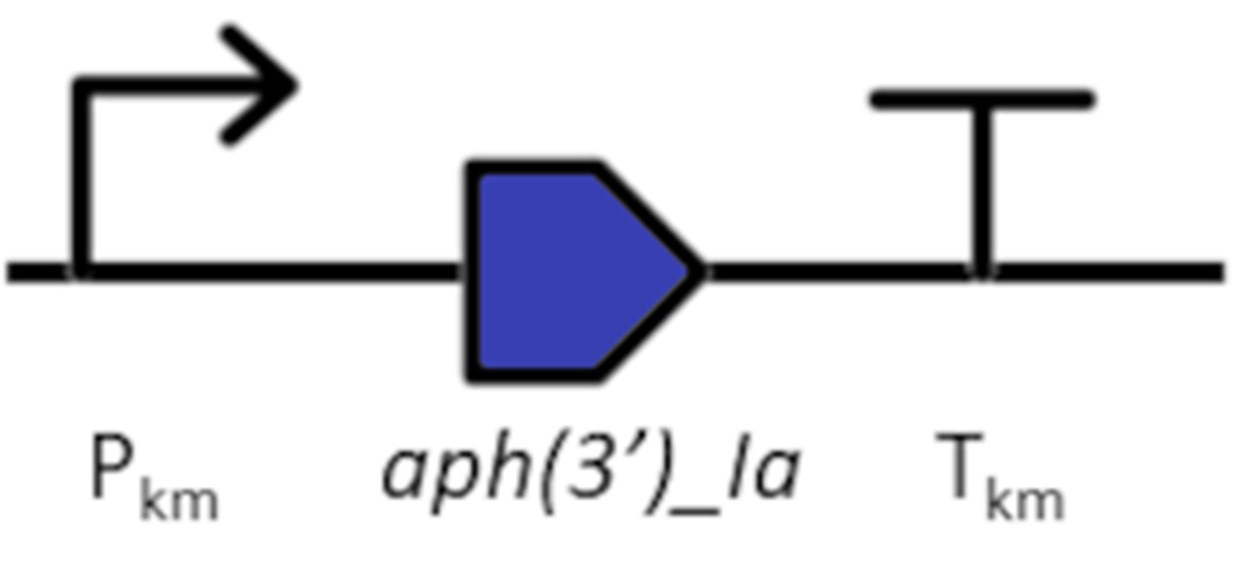
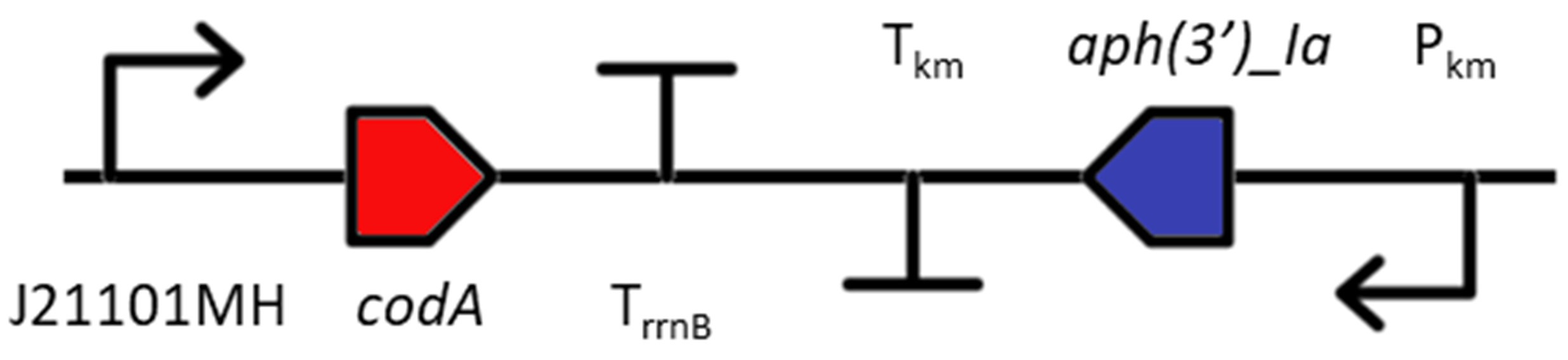
The KmR cassette was combined with a second marker for negative selection to allow users to further modify gene target sites (e.g. to generate an unmarked mutant). These consist of either the sacB gene from Bacillus subtilis encoding levansucrase (glycosyltransferase) (Lea-Smith et al., 2016 J Vis Exp 111: e54001) or a genetically modified cytosine deaminase (codA) gene (Young et al., 2014 Plant Journal 80: 915). The specific insertion sequence used for transformation (i.e. CodA-KmR or SacB-KmR) can be found under Search. Most Synechocystis sp. PCC 6803 marked mutants contain SacB-KmR.
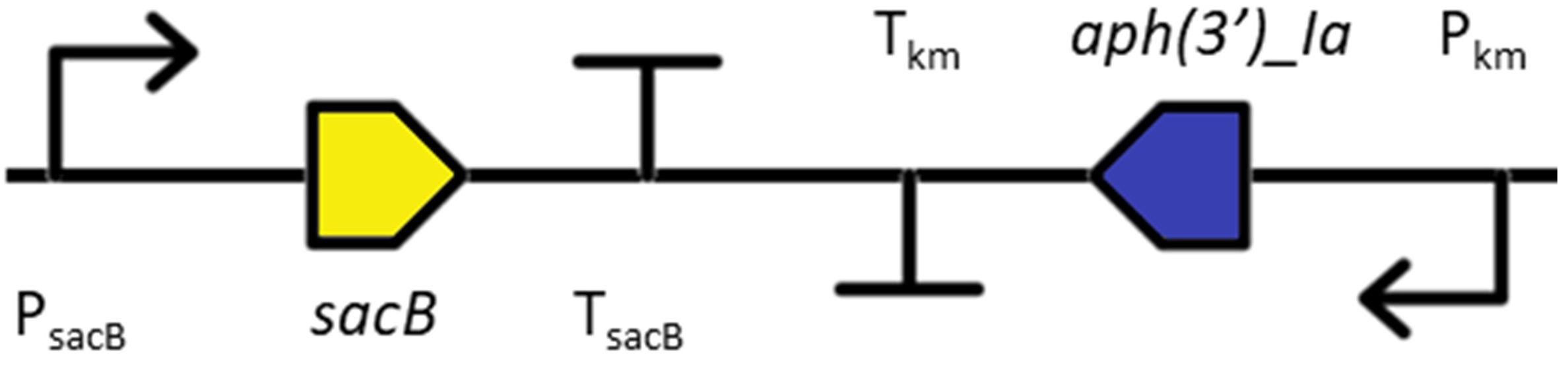
Outside of the insertion sequence are BpiI and AarI type IIS restriction sites that generate 4 bp overhangs. The sites closest to the Left Flank generate a TGCC overhang. The sites closest to the Right Flank generate a GGGA overhang. The TGCC-GGGA overhangs match those of the CyanoGate level T acceptor vector pPMQAK1-T (Vasudevan et al., 2019 Plant Physiol 180: 39), and are thus compatible with level 1 assemblies. Just inside of each flank region is a SapI type IIS restriction site that generates a 3 bp overhang. The site closest to the Left Flank generates a ATG overhang. The site closest to the Right Flank generate a TAA overhang. The SapI sites can be used for inserting a coding sequence in frame with native promoter and terminator sequences (Note: this is insertion specific, please check the flank sequences first).
Unmarking spacer plasmid
An unmarking spacer plasmid (pUC19K-UL) is available for modifying plasmids to remove and replace the insertion sequence with a short 63 bp spacer sequence (AATGAATACTGCTAAAACTGTTGTTGCAAAAAAATGAATTTAATCAAAAAGTTGTTCTCCTAG):
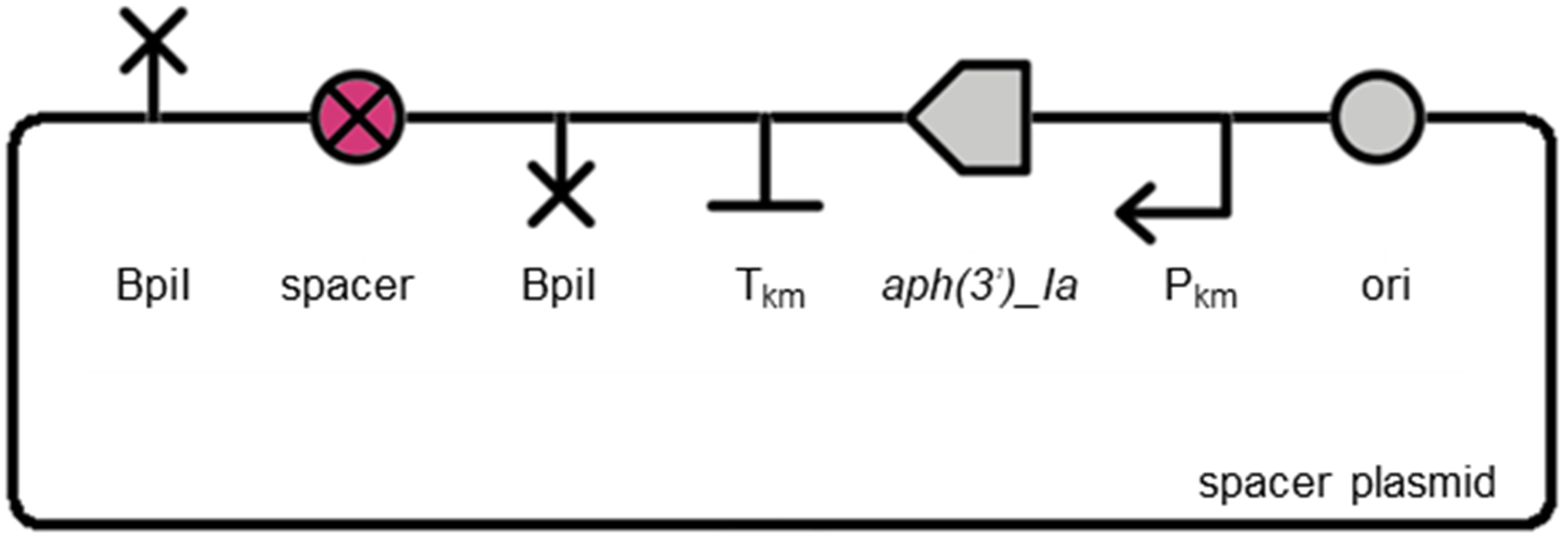
MoClo assembly of any gene specific KO plasmid (above) with pUC19K-UL using BpiI will result in the plasmid below, which can subsequently be used to unmarked marked Synechocystis sp. PCC 6803 mutants using appropriate counter-selection: